Extending Data Joins + Factors
Thursday, April 24
Today we will…
- Notes on Lab 3
- Project Info
- New Material
- Extensions to Data Joins
- Factors with
forcats - Clean Variable Names
- Lifecycle Stages
- Lab 4: Childcare Costs in California
Project
- Detailed information about the project is posted on Canvas
- You will complete the project in groups of 4
- The project is scaffolded into 5 “Checkpoints”
- The bulk of the work will be in Weeks 8-10
- First Checkpoint due 6th Tuesday (5/6) at 11:59pm
- Fill out a survey to form groups
- You can specify if there are people you want to work with or have me place you in a group
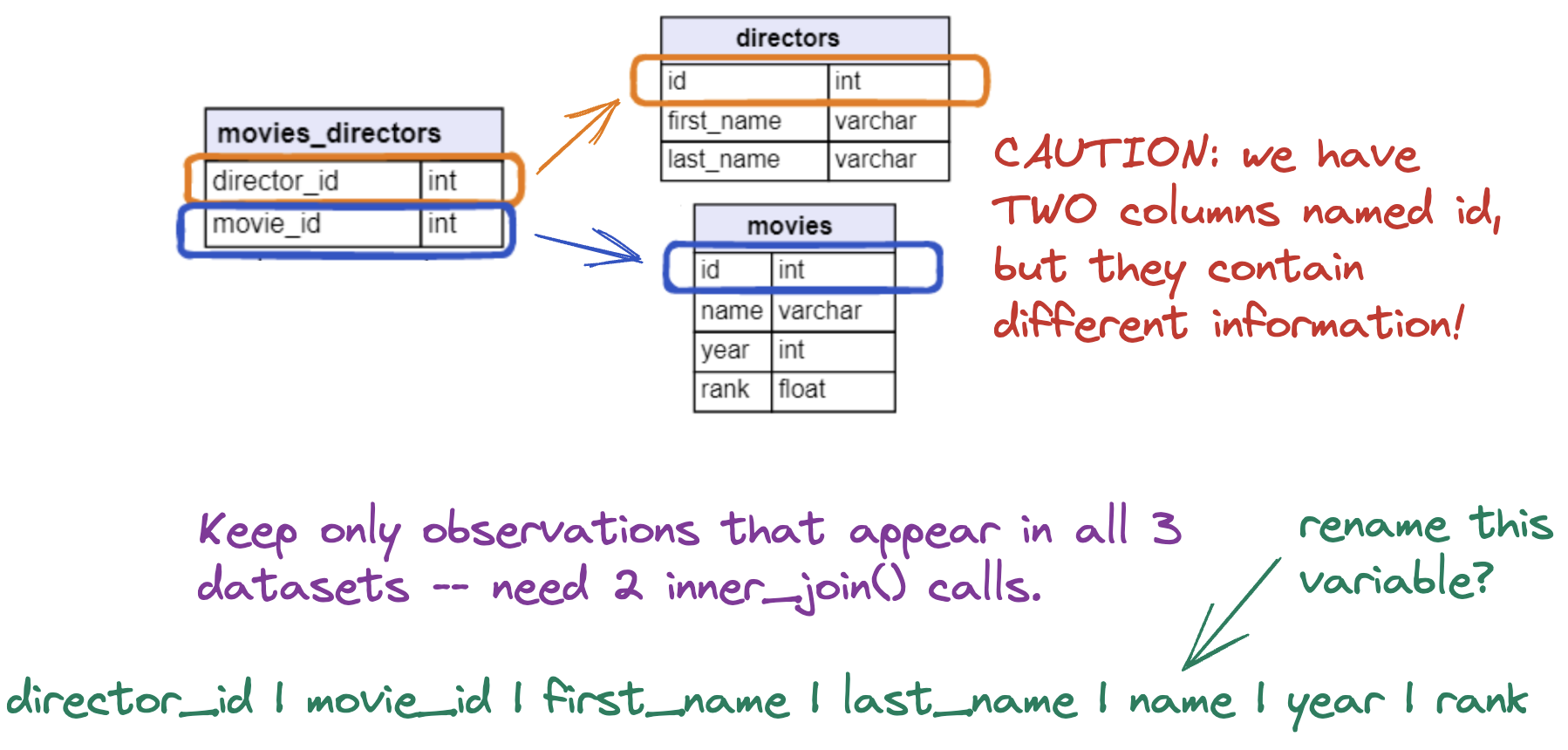
Extensions to Relational Data
Relational Data Reminder - Keys
When we work with relational data, we rely on keys.
- A key uniquely identifies an observation in a dataset.
- A key allows us to relate datasets to each other
IMDb Movies Data

What were the active years of each director?
Discussion
Which datasets do we need to use to answer this question?
Joining Multiple Data Sets
| director_id | movie_id |
|---|---|
| 429 | 300229 |
| 2931 | 254943 |
| 9247 | 124110 |
| 11652 | 10920 |
| director_id | movie_id | first_name | last_name |
|---|---|---|---|
| 429 | 300229 | Andrew | Adamson |
| 2931 | 254943 | Darren | Aronofsky |
| 9247 | 124110 | Zach | Braff |
| 11652 | 10920 | James (I) | Cameron |
| 11652 | 333856 | James (I) | Cameron |
| 14927 | 192017 | Ron | Clements |
| 15092 | 109093 | Ethan | Coen |
| 15092 | 237431 | Ethan | Coen |
| 15093 | 109093 | Joel | Coen |
| 15093 | 237431 | Joel | Coen |
| 15901 | 130128 | Francis Ford | Coppola |
| 15906 | 194874 | Sofia | Coppola |
| 16816 | 350424 | Cameron | Crowe |
| 17810 | 297838 | Frank | Darabont |
| 22104 | 224842 | Clint | Eastwood |
| 24758 | 112290 | David | Fincher |
| 28395 | 46169 | Mel (I) | Gibson |
| 35573 | 18979 | Ron | Howard |
| 35838 | 257264 | John (I) | Hughes |
| 37872 | 300229 | Vicky | Jenson |
| 38746 | 238695 | Mike (I) | Judge |
| 41975 | 314965 | David | Koepp |
| 44291 | 17173 | John (I) | Landis |
| 46315 | 344203 | Jay | Levey |
| 48115 | 313459 | George | Lucas |
| 56332 | 192017 | John | Musker |
| 58201 | 30959 | Christopher | Nolan |
| 58201 | 210511 | Christopher | Nolan |
| 65940 | 111813 | Rob | Reiner |
| 66849 | 306032 | Guy | Ritchie |
| 68161 | 116907 | Herbert (I) | Ross |
| 74758 | 238072 | Steven | Soderbergh |
| 76524 | 167324 | Oliver (I) | Stone |
| 78273 | 176711 | Quentin | Tarantino |
| 78273 | 176712 | Quentin | Tarantino |
| 78273 | 267038 | Quentin | Tarantino |
| 78273 | 276217 | Quentin | Tarantino |
| 82525 | 147603 | Paul (I) | Verhoeven |
| 83616 | 207992 | Andy | Wachowski |
| 83617 | 207992 | Larry | Wachowski |
| 88802 | 256630 | Unknown | Director |
| director_id | movie_id | first_name | last_name | name | year | rank |
|---|---|---|---|---|---|---|
| 429 | 300229 | Andrew | Adamson | Shrek | 2001 | 8.1 |
| 2931 | 254943 | Darren | Aronofsky | Pi | 1998 | 7.5 |
| 9247 | 124110 | Zach | Braff | Garden State | 2004 | 8.3 |
| 11652 | 10920 | James (I) | Cameron | Aliens | 1986 | 8.2 |
| 11652 | 333856 | James (I) | Cameron | Titanic | 1997 | 6.9 |
| 14927 | 192017 | Ron | Clements | Little Mermaid, The | 1989 | 7.3 |
| 15092 | 109093 | Ethan | Coen | Fargo | 1996 | 8.2 |
| 15092 | 237431 | Ethan | Coen | O Brother, Where Art Thou? | 2000 | 7.8 |
| 15093 | 109093 | Joel | Coen | Fargo | 1996 | 8.2 |
| 15093 | 237431 | Joel | Coen | O Brother, Where Art Thou? | 2000 | 7.8 |
| 15901 | 130128 | Francis Ford | Coppola | Godfather, The | 1972 | 9.0 |
| 15906 | 194874 | Sofia | Coppola | Lost in Translation | 2003 | 8.0 |
| 16816 | 350424 | Cameron | Crowe | Vanilla Sky | 2001 | 6.9 |
| 17810 | 297838 | Frank | Darabont | Shawshank Redemption, The | 1994 | 9.0 |
| 22104 | 224842 | Clint | Eastwood | Mystic River | 2003 | 8.1 |
| 24758 | 112290 | David | Fincher | Fight Club | 1999 | 8.5 |
| 28395 | 46169 | Mel (I) | Gibson | Braveheart | 1995 | 8.3 |
| 35573 | 18979 | Ron | Howard | Apollo 13 | 1995 | 7.5 |
| 35838 | 257264 | John (I) | Hughes | Planes, Trains & Automobiles | 1987 | 7.2 |
| 37872 | 300229 | Vicky | Jenson | Shrek | 2001 | 8.1 |
| 38746 | 238695 | Mike (I) | Judge | Office Space | 1999 | 7.6 |
| 41975 | 314965 | David | Koepp | Stir of Echoes | 1999 | 7.0 |
| 44291 | 17173 | John (I) | Landis | Animal House | 1978 | 7.5 |
| 46315 | 344203 | Jay | Levey | UHF | 1989 | 6.6 |
| 48115 | 313459 | George | Lucas | Star Wars | 1977 | 8.8 |
| 56332 | 192017 | John | Musker | Little Mermaid, The | 1989 | 7.3 |
| 58201 | 30959 | Christopher | Nolan | Batman Begins | 2005 | NA |
| 58201 | 210511 | Christopher | Nolan | Memento | 2000 | 8.7 |
| 65940 | 111813 | Rob | Reiner | Few Good Men, A | 1992 | 7.5 |
| 66849 | 306032 | Guy | Ritchie | Snatch. | 2000 | 7.9 |
| 68161 | 116907 | Herbert (I) | Ross | Footloose | 1984 | 5.8 |
| 74758 | 238072 | Steven | Soderbergh | Ocean's Eleven | 2001 | 7.5 |
| 76524 | 167324 | Oliver (I) | Stone | JFK | 1991 | 7.8 |
| 78273 | 176711 | Quentin | Tarantino | Kill Bill: Vol. 1 | 2003 | 8.4 |
| 78273 | 176712 | Quentin | Tarantino | Kill Bill: Vol. 2 | 2004 | 8.2 |
| 78273 | 267038 | Quentin | Tarantino | Pulp Fiction | 1994 | 8.7 |
| 78273 | 276217 | Quentin | Tarantino | Reservoir Dogs | 1992 | 8.3 |
| 82525 | 147603 | Paul (I) | Verhoeven | Hollow Man | 2000 | 5.3 |
| 83616 | 207992 | Andy | Wachowski | Matrix, The | 1999 | 8.5 |
| 83617 | 207992 | Larry | Wachowski | Matrix, The | 1999 | 8.5 |
| 88802 | 256630 | Unknown | Director | Pirates of the Caribbean | 2003 | NA |
| first_name | last_name | start_year | end_year | n_years_active |
|---|---|---|---|---|
| Quentin | Tarantino | 1992 | 2004 | 12 |
| James (I) | Cameron | 1986 | 1997 | 11 |
| Christopher | Nolan | 2000 | 2005 | 5 |
| Ethan | Coen | 1996 | 2000 | 4 |
| Joel | Coen | 1996 | 2000 | 4 |
| Andrew | Adamson | 2001 | 2001 | 0 |
| Andy | Wachowski | 1999 | 1999 | 0 |
| Cameron | Crowe | 2001 | 2001 | 0 |
| Clint | Eastwood | 2003 | 2003 | 0 |
| Darren | Aronofsky | 1998 | 1998 | 0 |
| David | Fincher | 1999 | 1999 | 0 |
| David | Koepp | 1999 | 1999 | 0 |
| Francis Ford | Coppola | 1972 | 1972 | 0 |
| Frank | Darabont | 1994 | 1994 | 0 |
| George | Lucas | 1977 | 1977 | 0 |
| Guy | Ritchie | 2000 | 2000 | 0 |
| Herbert (I) | Ross | 1984 | 1984 | 0 |
| Jay | Levey | 1989 | 1989 | 0 |
| John | Musker | 1989 | 1989 | 0 |
| John (I) | Hughes | 1987 | 1987 | 0 |
| John (I) | Landis | 1978 | 1978 | 0 |
| Larry | Wachowski | 1999 | 1999 | 0 |
| Mel (I) | Gibson | 1995 | 1995 | 0 |
| Mike (I) | Judge | 1999 | 1999 | 0 |
| Oliver (I) | Stone | 1991 | 1991 | 0 |
| Paul (I) | Verhoeven | 2000 | 2000 | 0 |
| Rob | Reiner | 1992 | 1992 | 0 |
| Ron | Clements | 1989 | 1989 | 0 |
| Ron | Howard | 1995 | 1995 | 0 |
| Sofia | Coppola | 2003 | 2003 | 0 |
| Steven | Soderbergh | 2001 | 2001 | 0 |
| Unknown | Director | 2003 | 2003 | 0 |
| Vicky | Jenson | 2001 | 2001 | 0 |
| Zach | Braff | 2004 | 2004 | 0 |
Know Your Unique Observations!
Discussion
What is the observational unit after joining the directors and movies_directors by the director_id key? What happens for directors that have multiple movies in the movies_directors data?
| id | first_name | last_name | movie_id |
|---|---|---|---|
| 429 | Andrew | Adamson | 300229 |
| 2931 | Darren | Aronofsky | 254943 |
| 9247 | Zach | Braff | 124110 |
| 11652 | James (I) | Cameron | 10920 |
| 11652 | James (I) | Cameron | 333856 |
| 14927 | Ron | Clements | 192017 |
| 15092 | Ethan | Coen | 109093 |
| 15092 | Ethan | Coen | 237431 |
| 15093 | Joel | Coen | 109093 |
| 15093 | Joel | Coen | 237431 |
| 15901 | Francis Ford | Coppola | 130128 |
| 15906 | Sofia | Coppola | 194874 |
| 16816 | Cameron | Crowe | 350424 |
| 17810 | Frank | Darabont | 297838 |
| 22104 | Clint | Eastwood | 224842 |
| 24758 | David | Fincher | 112290 |
| 28395 | Mel (I) | Gibson | 46169 |
| 35573 | Ron | Howard | 18979 |
| 35838 | John (I) | Hughes | 257264 |
| 37872 | Vicky | Jenson | 300229 |
| 38746 | Mike (I) | Judge | 238695 |
| 41975 | David | Koepp | 314965 |
| 44291 | John (I) | Landis | 17173 |
| 46315 | Jay | Levey | 344203 |
| 48115 | George | Lucas | 313459 |
| 56332 | John | Musker | 192017 |
| 58201 | Christopher | Nolan | 30959 |
| 58201 | Christopher | Nolan | 210511 |
| 65940 | Rob | Reiner | 111813 |
| 66849 | Guy | Ritchie | 306032 |
| 68161 | Herbert (I) | Ross | 116907 |
| 74758 | Steven | Soderbergh | 238072 |
| 76524 | Oliver (I) | Stone | 167324 |
| 78273 | Quentin | Tarantino | 176711 |
| 78273 | Quentin | Tarantino | 176712 |
| 78273 | Quentin | Tarantino | 267038 |
| 78273 | Quentin | Tarantino | 276217 |
| 82525 | Paul (I) | Verhoeven | 147603 |
| 83616 | Andy | Wachowski | 207992 |
| 83617 | Larry | Wachowski | 207992 |
| 88802 | Unknown | Director | 256630 |
Know Your Unique Observations!
Remember the rodent data from Lab 2. Say we had separate datsets for measurements and species information:
| genus | species | taxa | species_id |
|---|---|---|---|
| Dipodomys | merriami | Rodent | DM |
| Dipodomys | ordii | Rodent | DO |
| Perognathus | flavus | Rodent | PF |
| Chaetodipus | penicillatus | Rodent | PP |
| Peromyscus | eremicus | Rodent | PE |
| Onychomys | leucogaster | Rodent | OL |
| Reithrodontomys | megalotis | Rodent | RM |
| Dipodomys | spectabilis | Rodent | DS |
| Onychomys | torridus | Rodent | OT |
| Neotoma | albigula | Rodent | NL |
| Peromyscus | maniculatus | Rodent | PM |
| Sigmodon | hispidus | Rodent | SH |
| Reithrodontomys | fulvescens | Rodent | RF |
| Chaetodipus | baileyi | Rodent | PB |
| genus_name | species | sex | hindfoot_length | weight |
|---|---|---|---|---|
| Dipodomys | merriami | M | 35 | 40 |
| Dipodomys | merriami | M | 37 | 48 |
| Dipodomys | merriami | F | 34 | 29 |
| Dipodomys | merriami | F | 35 | 46 |
| Dipodomys | merriami | M | 35 | 36 |
| Dipodomys | ordii | F | 32 | 52 |
| Perognathus | flavus | M | 15 | 8 |
| Dipodomys | merriami | F | 36 | 35 |
| Perognathus | flavus | M | 12 | 7 |
| Dipodomys | merriami | F | 32 | 22 |
| Perognathus | flavus | M | 16 | 9 |
| Dipodomys | merriami | F | 34 | 42 |
| Perognathus | flavus | F | 14 | 8 |
| Dipodomys | merriami | F | 35 | 41 |
| Dipodomys | merriami | F | 37 | 37 |
| Dipodomys | merriami | F | 35 | 43 |
| Dipodomys | merriami | F | 35 | 41 |
| Dipodomys | merriami | F | 33 | 40 |
| Perognathus | flavus | F | 11 | 9 |
| Dipodomys | merriami | F | 35 | 45 |
| Chaetodipus | penicillatus | F | 20 | 15 |
| Dipodomys | merriami | M | 35 | 29 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | F | 36 | 43 |
| Dipodomys | merriami | M | 38 | 46 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 38 | 40 |
| Dipodomys | merriami | M | 37 | 45 |
| Dipodomys | merriami | F | 35 | 46 |
| Dipodomys | merriami | F | 35 | 40 |
| Dipodomys | merriami | F | 35 | 30 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | M | 35 | 34 |
| Dipodomys | merriami | F | 37 | 42 |
| Dipodomys | merriami | M | 37 | 42 |
| Perognathus | flavus | F | 13 | 8 |
| Dipodomys | merriami | F | 37 | 31 |
| Dipodomys | merriami | F | 36 | 40 |
| Dipodomys | merriami | M | 36 | 37 |
| Dipodomys | merriami | M | 36 | 48 |
| Dipodomys | merriami | M | 37 | 42 |
| Dipodomys | merriami | F | 39 | 45 |
| Chaetodipus | penicillatus | F | 21 | 16 |
| Dipodomys | merriami | F | 36 | 36 |
| Dipodomys | merriami | M | 36 | 42 |
| Dipodomys | merriami | M | 36 | 44 |
| Dipodomys | merriami | F | 36 | 41 |
| Dipodomys | merriami | F | 36 | 40 |
| Dipodomys | merriami | M | 37 | 34 |
| Dipodomys | merriami | M | 33 | 40 |
| Dipodomys | merriami | M | 33 | 44 |
| Dipodomys | merriami | M | 37 | 44 |
| Dipodomys | merriami | M | 34 | 36 |
| Dipodomys | merriami | M | 35 | 33 |
| Dipodomys | merriami | F | 37 | 46 |
| Dipodomys | merriami | F | 34 | 35 |
| Dipodomys | merriami | M | 36 | 46 |
| Dipodomys | merriami | F | 33 | 37 |
| Dipodomys | merriami | M | 36 | 34 |
| Dipodomys | merriami | F | 36 | 45 |
| Perognathus | flavus | F | 15 | 7 |
| Dipodomys | merriami | M | 37 | 51 |
| Dipodomys | merriami | M | 35 | 39 |
| Dipodomys | merriami | M | 36 | 29 |
| Dipodomys | merriami | F | 32 | 48 |
| Dipodomys | merriami | M | 38 | 46 |
| Dipodomys | merriami | F | 37 | 41 |
| Dipodomys | merriami | M | 37 | 45 |
| Dipodomys | merriami | F | 35 | 42 |
| Dipodomys | merriami | F | 36 | 53 |
| Dipodomys | merriami | F | 35 | 49 |
| Dipodomys | merriami | F | 36 | 46 |
| Perognathus | flavus | F | 13 | 9 |
| Chaetodipus | penicillatus | F | 19 | 15 |
| Perognathus | flavus | M | 13 | 4 |
| Dipodomys | merriami | M | 36 | 48 |
| Dipodomys | merriami | M | 37 | 51 |
| Dipodomys | merriami | M | 38 | 50 |
| Dipodomys | merriami | M | 35 | 44 |
| Dipodomys | merriami | M | 25 | 44 |
| Dipodomys | merriami | M | 35 | 45 |
| Dipodomys | merriami | F | 37 | 45 |
| Peromyscus | eremicus | M | 20 | 19 |
| Dipodomys | merriami | F | 38 | 44 |
| Dipodomys | merriami | F | 36 | 42 |
| Dipodomys | merriami | M | 37 | 39 |
| Dipodomys | merriami | M | 37 | 47 |
| Dipodomys | merriami | M | 36 | 42 |
| Dipodomys | merriami | M | 36 | 49 |
| Dipodomys | merriami | M | 38 | 39 |
| Dipodomys | merriami | F | 36 | 43 |
| Dipodomys | merriami | M | 35 | 50 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | M | 37 | 47 |
| Dipodomys | merriami | F | 36 | 37 |
| Dipodomys | merriami | M | 36 | 41 |
| Dipodomys | merriami | F | 36 | 36 |
| Dipodomys | merriami | M | 36 | 45 |
| Peromyscus | eremicus | M | 19 | 20 |
Know Your Unique Observations!
Discussion
What happens if we join species and measurements by the genus only?
| genus_name | species.x | sex | hindfoot_length | weight | species.y | taxa | species_id |
|---|---|---|---|---|---|---|---|
| Dipodomys | merriami | M | 35 | 40 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 40 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 40 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 37 | 48 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 37 | 48 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 37 | 48 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 34 | 29 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 34 | 29 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 34 | 29 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 46 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 46 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 46 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 35 | 36 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 36 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 36 | spectabilis | Rodent | DS |
| Dipodomys | ordii | F | 32 | 52 | merriami | Rodent | DM |
| Dipodomys | ordii | F | 32 | 52 | ordii | Rodent | DO |
| Dipodomys | ordii | F | 32 | 52 | spectabilis | Rodent | DS |
| Perognathus | flavus | M | 15 | 8 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 36 | 35 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 36 | 35 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 36 | 35 | spectabilis | Rodent | DS |
| Perognathus | flavus | M | 12 | 7 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 32 | 22 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 32 | 22 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 32 | 22 | spectabilis | Rodent | DS |
| Perognathus | flavus | M | 16 | 9 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 34 | 42 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 34 | 42 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 34 | 42 | spectabilis | Rodent | DS |
| Perognathus | flavus | F | 14 | 8 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 35 | 41 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 41 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 41 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 37 | 37 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 37 | 37 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 37 | 37 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 43 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 43 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 43 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 41 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 41 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 41 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 33 | 40 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 33 | 40 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 33 | 40 | spectabilis | Rodent | DS |
| Perognathus | flavus | F | 11 | 9 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 35 | 45 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 45 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 45 | spectabilis | Rodent | DS |
| Chaetodipus | penicillatus | F | 20 | 15 | penicillatus | Rodent | PP |
| Chaetodipus | penicillatus | F | 20 | 15 | baileyi | Rodent | PB |
| Dipodomys | merriami | M | 35 | 29 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 29 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 29 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 35 | 39 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 39 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 39 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 36 | 43 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 36 | 43 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 36 | 43 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 38 | 46 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 38 | 46 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 38 | 46 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 36 | 41 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 36 | 41 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 36 | 41 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 36 | 41 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 36 | 41 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 38 | 40 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 38 | 40 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 38 | 40 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 37 | 45 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 37 | 45 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 37 | 45 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 46 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 46 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 46 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 40 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 40 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 40 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 35 | 30 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 35 | 30 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 35 | 30 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 35 | 39 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 39 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 39 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 35 | 34 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 35 | 34 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 35 | 34 | spectabilis | Rodent | DS |
| Dipodomys | merriami | F | 37 | 42 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 37 | 42 | ordii | Rodent | DO |
| Dipodomys | merriami | F | 37 | 42 | spectabilis | Rodent | DS |
| Dipodomys | merriami | M | 37 | 42 | merriami | Rodent | DM |
| Dipodomys | merriami | M | 37 | 42 | ordii | Rodent | DO |
| Dipodomys | merriami | M | 37 | 42 | spectabilis | Rodent | DS |
| Perognathus | flavus | F | 13 | 8 | flavus | Rodent | PF |
| Dipodomys | merriami | F | 37 | 31 | merriami | Rodent | DM |
| Dipodomys | merriami | F | 37 | 31 | ordii | Rodent | DO |
DANGER : MANY-TO-MANY JOIN!
Our observations exploded and the species_id isn’t even right for some observations! We also now have a species.x and species.y variable since the variable was present in both the left and right data.
Joining on Multiple Variables
To fix this, we need to join on multiple variables (a compound key):
| genus | species | taxa | species_id | sex | hindfoot_length | weight |
|---|---|---|---|---|---|---|
| Dipodomys | merriami | Rodent | DM | M | 35 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 48 |
| Dipodomys | merriami | Rodent | DM | F | 34 | 29 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 46 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 36 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 35 |
| Dipodomys | merriami | Rodent | DM | F | 32 | 22 |
| Dipodomys | merriami | Rodent | DM | F | 34 | 42 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 41 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 37 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 43 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 41 |
| Dipodomys | merriami | Rodent | DM | F | 33 | 40 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 45 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 29 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 39 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 43 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 46 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 45 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 46 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 40 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 30 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 39 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 34 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 42 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 42 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 31 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 37 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 48 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 42 |
| Dipodomys | merriami | Rodent | DM | F | 39 | 45 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 36 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 42 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 44 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 41 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 34 |
| Dipodomys | merriami | Rodent | DM | M | 33 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 33 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 34 | 36 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 33 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 46 |
| Dipodomys | merriami | Rodent | DM | F | 34 | 35 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 46 |
| Dipodomys | merriami | Rodent | DM | F | 33 | 37 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 34 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 45 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 51 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 39 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 29 |
| Dipodomys | merriami | Rodent | DM | F | 32 | 48 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 46 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 45 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 42 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 53 |
| Dipodomys | merriami | Rodent | DM | F | 35 | 49 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 46 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 48 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 51 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 50 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 25 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 45 |
| Dipodomys | merriami | Rodent | DM | F | 37 | 45 |
| Dipodomys | merriami | Rodent | DM | F | 38 | 44 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 42 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 39 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 47 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 42 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 49 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 39 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 43 |
| Dipodomys | merriami | Rodent | DM | M | 35 | 50 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 47 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 37 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 41 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 36 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 45 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 40 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 49 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 55 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 46 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 38 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 46 |
| Dipodomys | merriami | Rodent | DM | M | 34 | 36 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 44 |
| Dipodomys | merriami | Rodent | DM | M | 36 | 53 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 41 |
| Dipodomys | merriami | Rodent | DM | M | 37 | 48 |
| Dipodomys | merriami | Rodent | DM | M | 38 | 47 |
| Dipodomys | merriami | Rodent | DM | F | 36 | 42 |
Factor Variables
What is a factor variable?
Factors are used for
- categorical variables with a fixed and known set of possible values.
- E.g.,
day_born= Sunday, Monday, Tuesday, …, Saturday
- displaying character vectors in non-alphabetical order.
- useful for nice tables and plots!
Eras Tour
Let’s consider songs that Taylor Swift played on her Eras Tour.
I have randomly selected 25 songs (and their albums) to demonstrate.
| Song | Album |
|---|---|
| 22 | Red |
| ...Ready for It? | Reputation |
| The Archer | Lover |
| Bejeweled | Midnights |
| Style | 1989 |
| You Belong With Me | Fearless |
| Don't Blame Me | Reputation |
| illicit affairs | Folklore |
| Lavender Haze | Midnights |
| marjorie | Evermore |
Creating a Factor – Base R
A character vector:
[1] "Red" "Reputation" "Lover" "Midnights" "1989"
[6] "Fearless" "Reputation" "Folklore" "Midnights" "Evermore"
[11] "Evermore" "Lover" "Lover" "Red" "Reputation"
[16] "Reputation" "Speak Now" "Red" "Midnights" "Fearless"
[21] "1989" "Midnights" "Fearless" "Folklore" "Lover" A factor vector:
[1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: 1989 Evermore Fearless Folklore Lover Midnights Red ... Speak NowCreating a Factor – Base R
When you create a factor variable from a vector…
- Every unique element in the vector becomes a level.
- The levels are ordered alphabetically.
- The elements are no longer displayed in quotes.
Creating a Factor – Base R
You can specify the order of the levels with the level argument.
eras_data |>
pull(Album) |>
factor(levels = c("Fearless","Speak Now","Red","1989",
"Reputation","Lover","Folklore",
"Evermore","Midnights")) [1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: Fearless Speak Now Red 1989 Reputation Lover Folklore ... Midnightsforcats
We use this package to…
turn character variables into factors.
make factors by discretizing numeric variables.
rename or reorder the levels of an existing factor.

Note
The packages forcats (“for categoricals”) helps wrangle categorical variables.
forcatsloads withtidyverse!
Creating a Factor – fct
With fct(), the levels are automatically ordered in the order of first appearance.
[1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: Red Reputation Lover Midnights 1989 Fearless Folklore ... Speak NowCreating a Factor – fct
You can still specify the order of the levels with level.
eras_data |>
mutate(Album = fct(Album,
levels = c("Fearless","Speak Now","Red",
"1989", "Reputation","Lover",
"Folklore", "Evermore","Midnights"))) |>
pull(Album) [1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
9 Levels: Fearless Speak Now Red 1989 Reputation Lover Folklore ... MidnightsCreating a Factor – fct
You can also specify non-present levels.
eras_data |>
mutate(Album = fct(Album,
levels = c("Taylor Swift",
"Fearless","Speak Now","Red",
"1989", "Reputation","Lover",
"Folklore", "Evermore","Midnights",
"The Tortured Poets Department"))) |>
pull(Album) [1] Red Reputation Lover Midnights 1989 Fearless
[7] Reputation Folklore Midnights Evermore Evermore Lover
[13] Lover Red Reputation Reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless Folklore
[25] Lover
11 Levels: Taylor Swift Fearless Speak Now Red 1989 Reputation ... The Tortured Poets DepartmentRe-coding a Factor – fct_recode
Oops, we have a typo in some of our levels! We change existing levels with the syntax <new level> = <old level>.
eras_data |>
mutate(Album = fct_recode(.f = Album,
"folklore" = "Folklore",
"evermore" = "Evermore",
"reputation" = "Reputation")) |>
pull(Album) [1] Red reputation Lover Midnights 1989 Fearless
[7] reputation folklore Midnights evermore evermore Lover
[13] Lover Red reputation reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless folklore
[25] Lover
11 Levels: Taylor Swift Fearless Speak Now Red 1989 reputation ... The Tortured Poets DepartmentNon-specified levels are not re-coded.
Re-coding a Factor – case_when
We have similar functionality with the case_when() function…
eras_data |>
mutate(Album = case_when(Album == "Folklore" ~ "folklore",
Album == "Evermore" ~ "evermore",
Album == "Reputation" ~ "reputation",
.default = Album),
Album = fct(Album)) |>
pull(Album) [1] Red reputation Lover Midnights 1989 Fearless
[7] reputation folklore Midnights evermore evermore Lover
[13] Lover Red reputation reputation Speak Now Red
[19] Midnights Fearless 1989 Midnights Fearless folklore
[25] Lover
9 Levels: Red reputation Lover Midnights 1989 Fearless folklore ... Speak NowCollapsing a Factor –fct_collapse
Collapse multiple existing levels of a factor with the syntax <new level> = c(<old levels>).
| Song | Album | Genre |
|---|---|---|
| willow | evermore | folk pop |
| You Belong With Me | Fearless | country pop |
| Lavender Haze | Midnights | alt-pop |
| We Are Never Ever Getting Back Together | Red | pop rock |
| illicit affairs | folklore | folk pop |
| Look What You Made Me Do | reputation | electropop |
Re-leveling a Factor –fct_relevel
Change the order of the levels of an existing factor.
eras_data |>
mutate(Album = fct_relevel(.f = Album,
c("Fearless","1989","Taylor Swift",
"Speak Now","Red","Midnights","reputation",
"folklore","Lover","evermore"))) |>
pull(Album) |>
levels() [1] "Fearless" "1989"
[3] "Taylor Swift" "Speak Now"
[5] "Red" "Midnights"
[7] "reputation" "folklore"
[9] "Lover" "evermore"
[11] "The Tortured Poets Department"Unspecified levels remain in the same order at the end.
Re-ordering Factors in ggplot2
The bars follow the default factor levels.
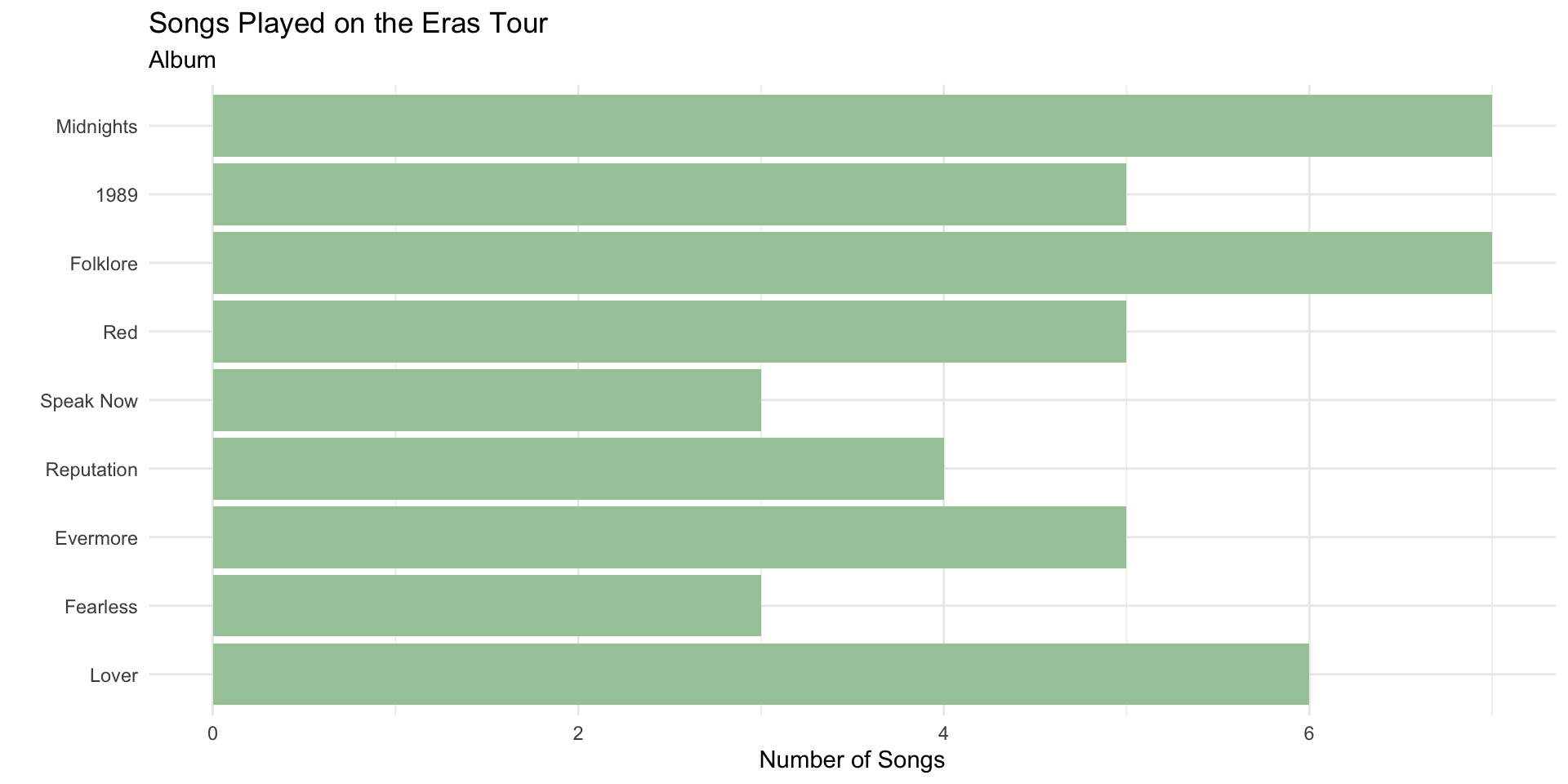
We can order factor levels to order the bar plot.
full_eras |>
mutate(Album = fct(Album,
levels = c("Fearless","Speak Now","Red",
"1989","Reputation","Lover",
"Folklore","Evermore",
"Midnights"))) |>
ggplot() +
geom_bar(aes(y = Album), fill = "#A5C9A5") +
theme_minimal() +
labs(x = "Number of Songs",
y = "",
subtitle = "Album",
title = "Songs Played on the Eras Tour")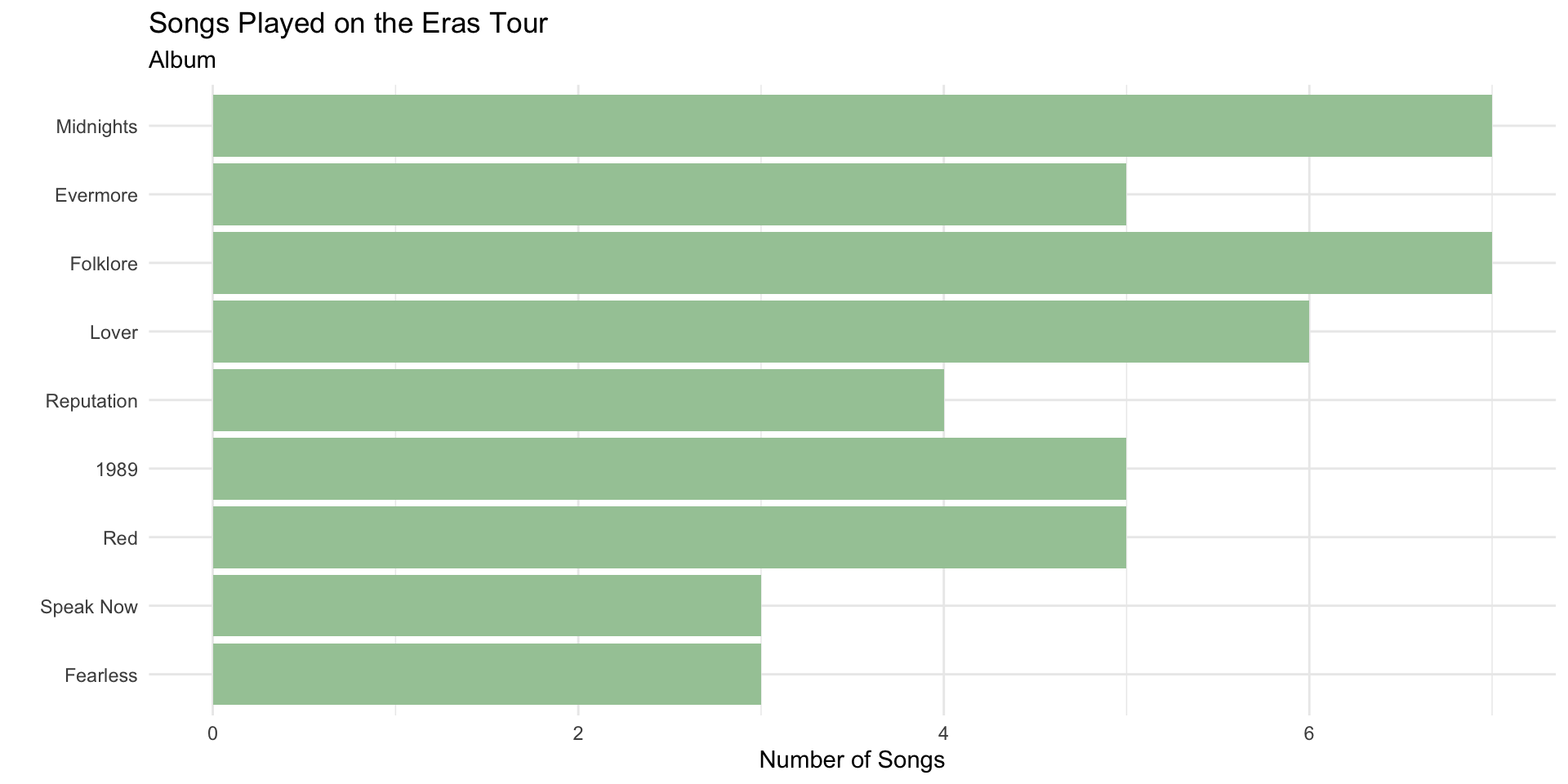
Re-ordering Factors in ggplot2
The bars follow the default factor levels.
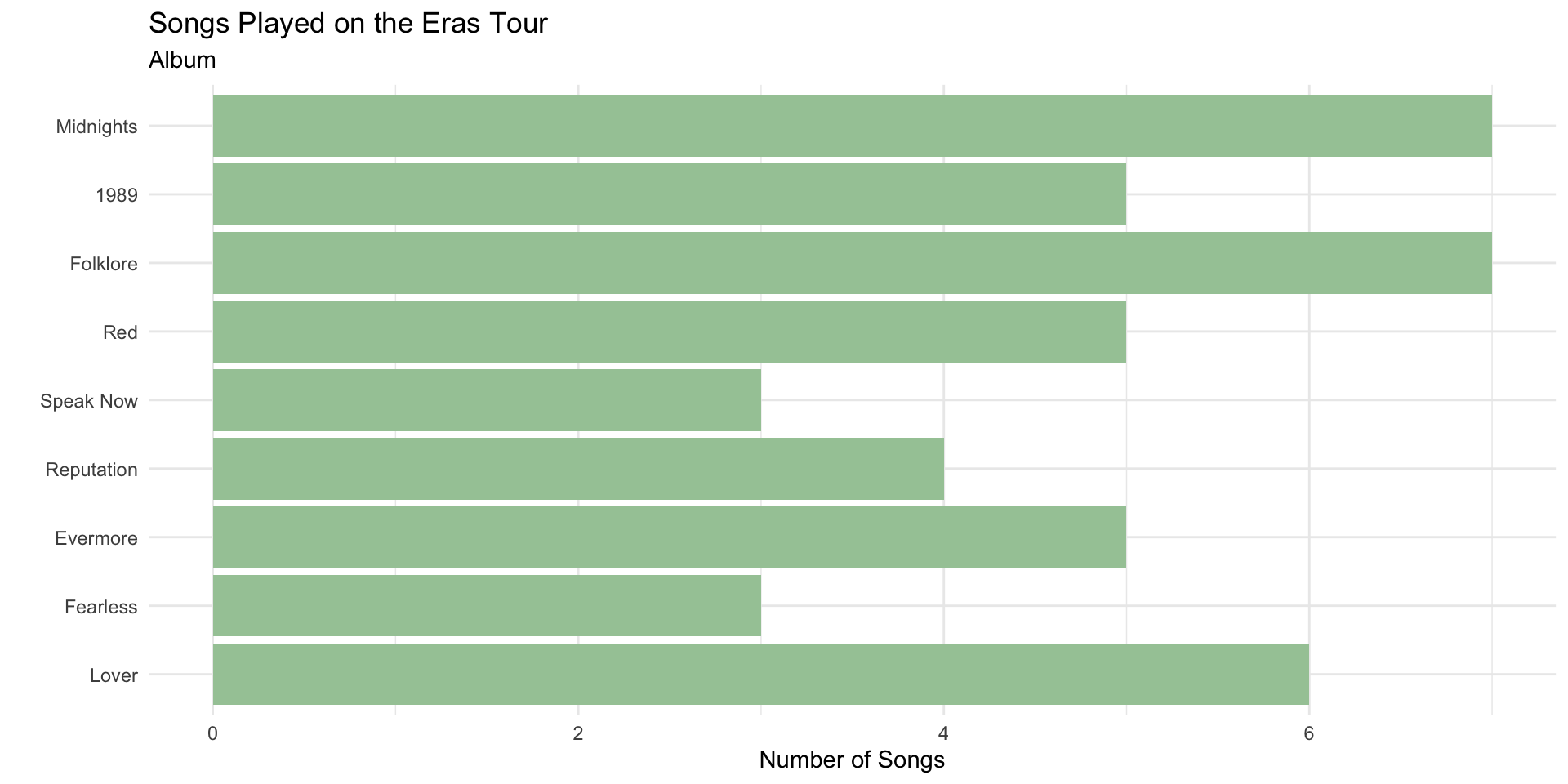
We can order factor levels to order the bar plot by the count using fct_infreq()
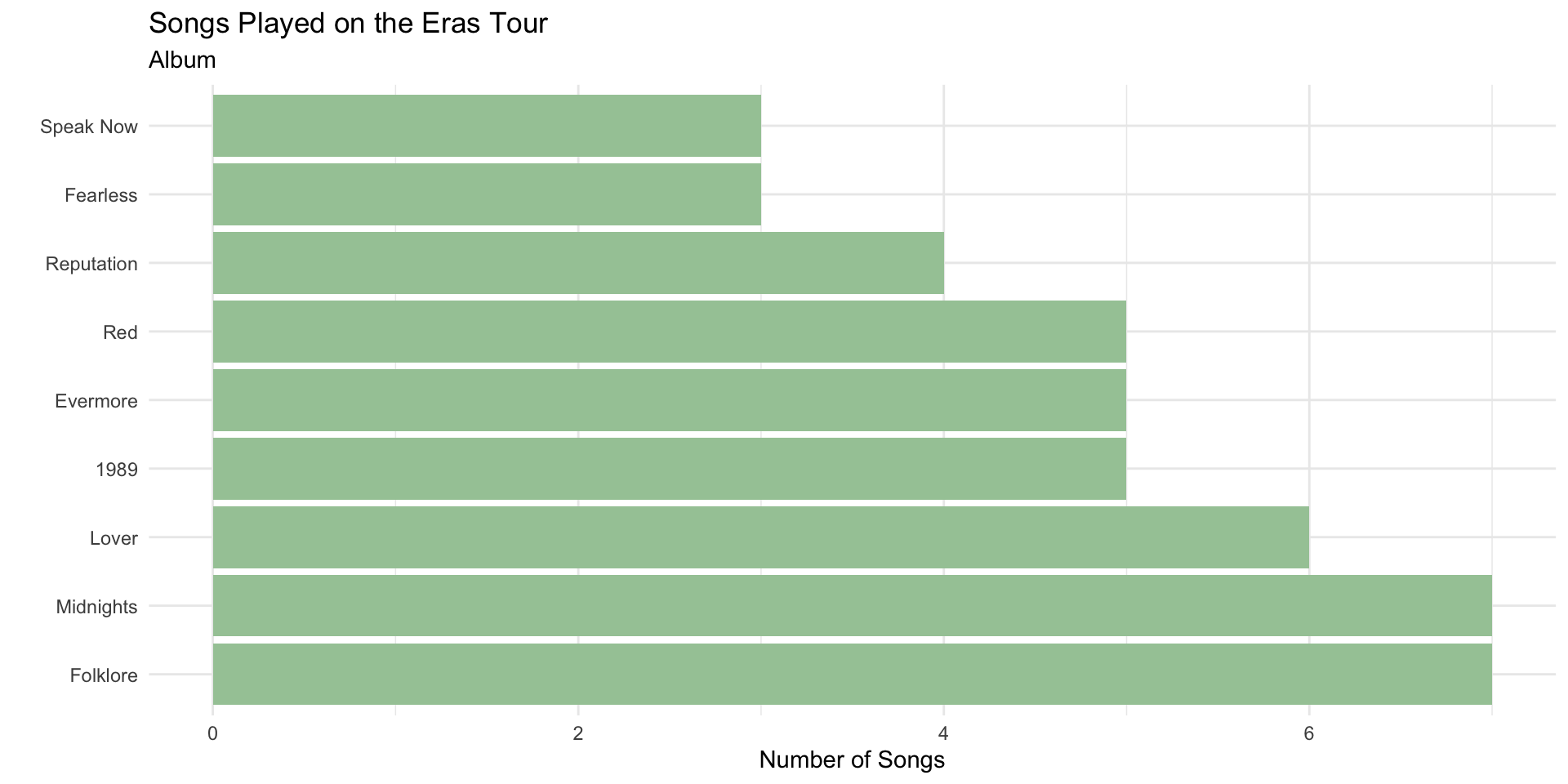
Re-ordering Factors in ggplot2
The ridge plots follow the order of the factor levels.
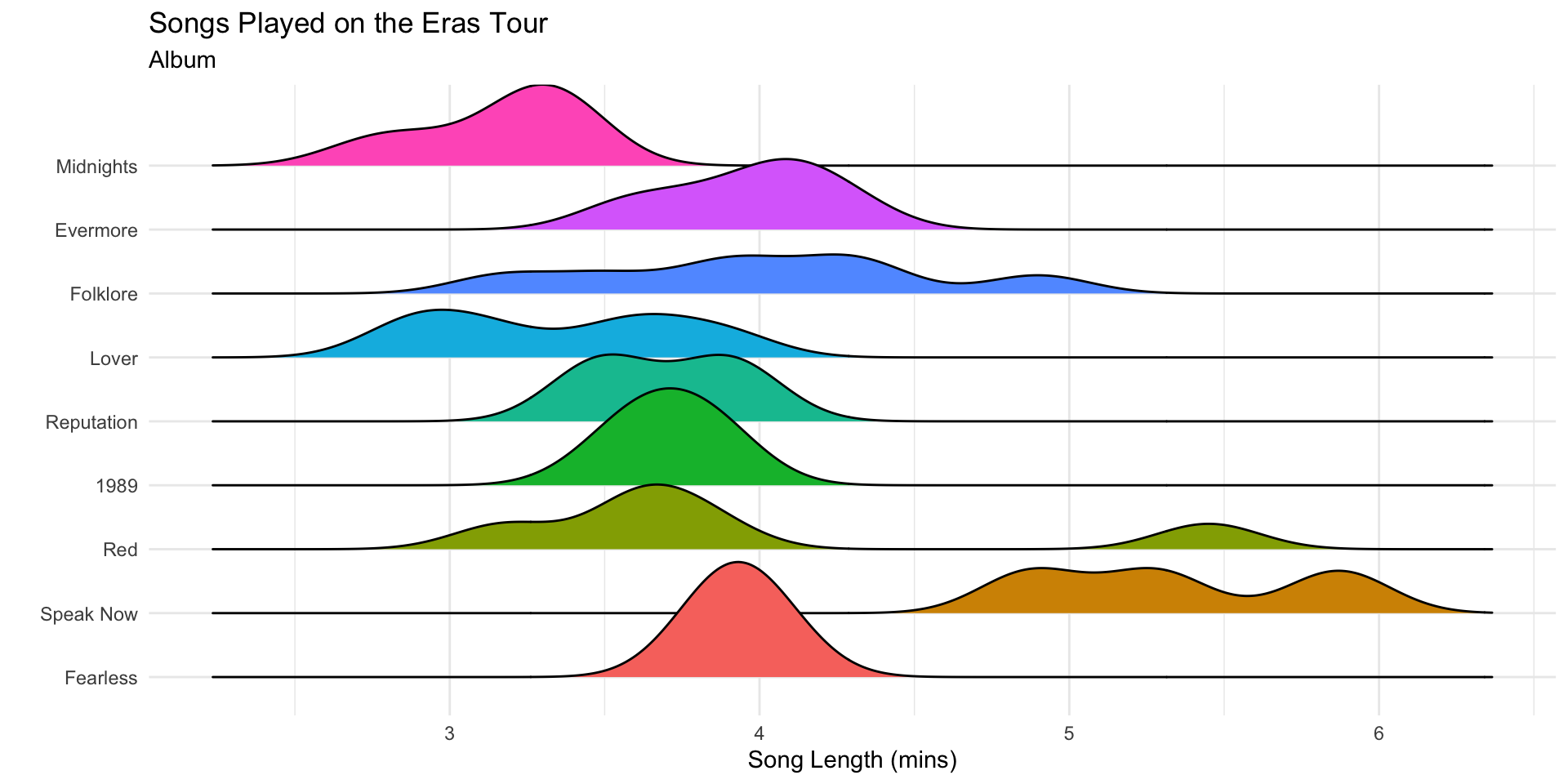
Inside ggplot(), we can order factor levels by a summary value.
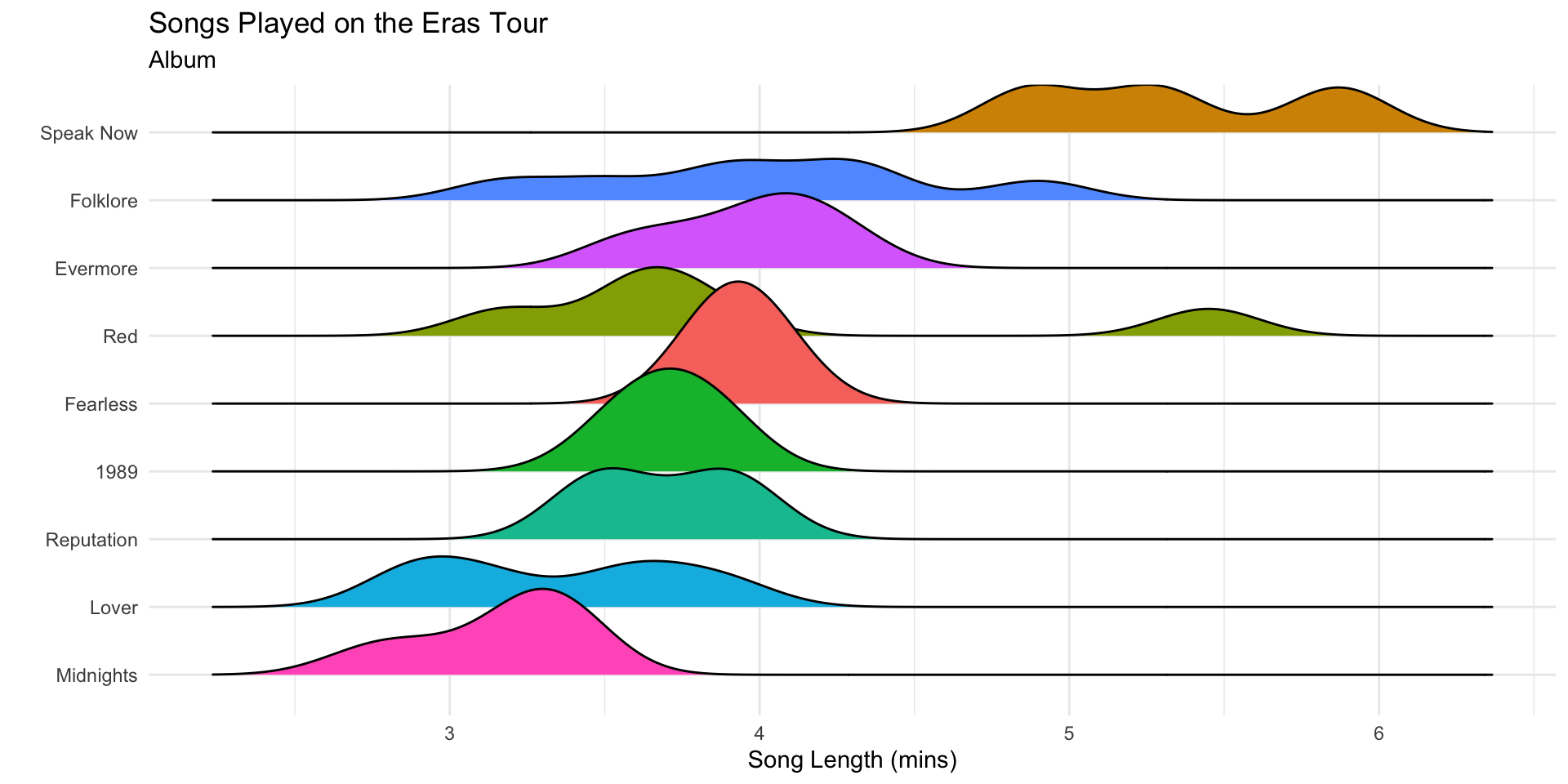
Re-ordering Factors in ggplot2
Remember the miliary data from the practice activity?
The legend follows the order of the factor levels.
military_long |>
filter(Country %in% central.asia,
!is.na(spending)) |>
ggplot(aes(x = year,
y = spending,
color = Country)) +
geom_line() +
labs(x = "Year",
y = "",
subtitle = "Spending (as % of Government Spending)",
title = "Military Expenditures in Central Asia") +
scale_color_manual(values = brewer.pal(3, "Dark2")) +
scale_y_continuous(labels = scales::percent) +
scale_x_continuous(breaks = seq(1990, 2023, 5)) +
theme_bw() +
theme(panel.grid.minor.x = element_blank())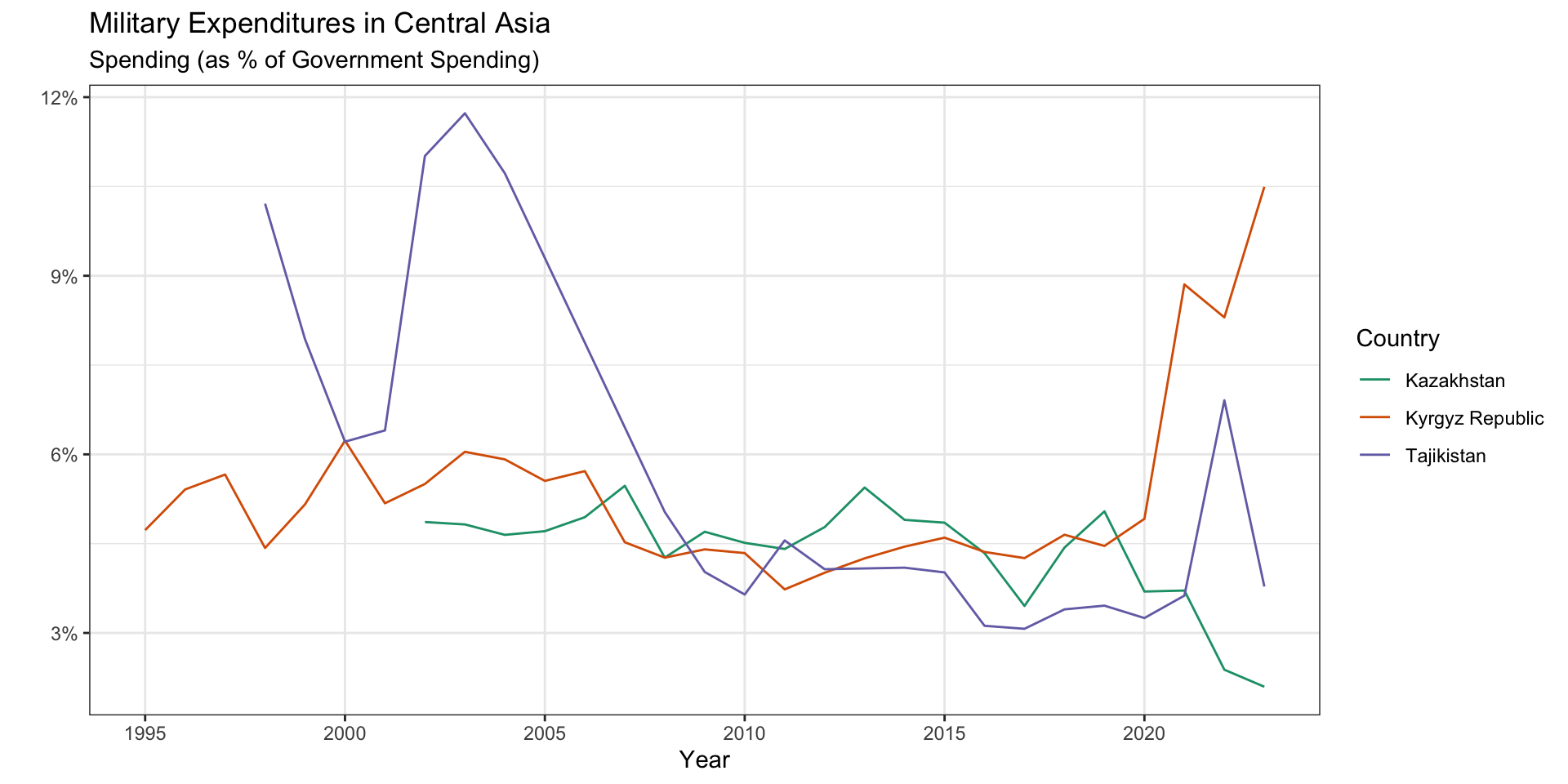
Inside ggplot(), we can order factor levels by the \(y\) values associated with the largest \(x\) values.
military_long |>
filter(Country %in% central.asia,
!is.na(spending)) |>
ggplot(aes(x = year,
y = spending,
color = fct_reorder2(.x = year,
.y = spending,
.f = Country))) +
geom_line() +
labs(x = "Year",
y = "",
color = "Country",
subtitle = "Spending (as % of Government Spending)",
title = "Military Expenditures in Central Asia") +
scale_color_manual(values = brewer.pal(3, "Dark2")) +
scale_y_continuous(labels = scales::percent) +
scale_x_continuous(breaks = seq(1990, 2023, 5)) +
theme_bw() +
theme(panel.grid.minor.x = element_blank())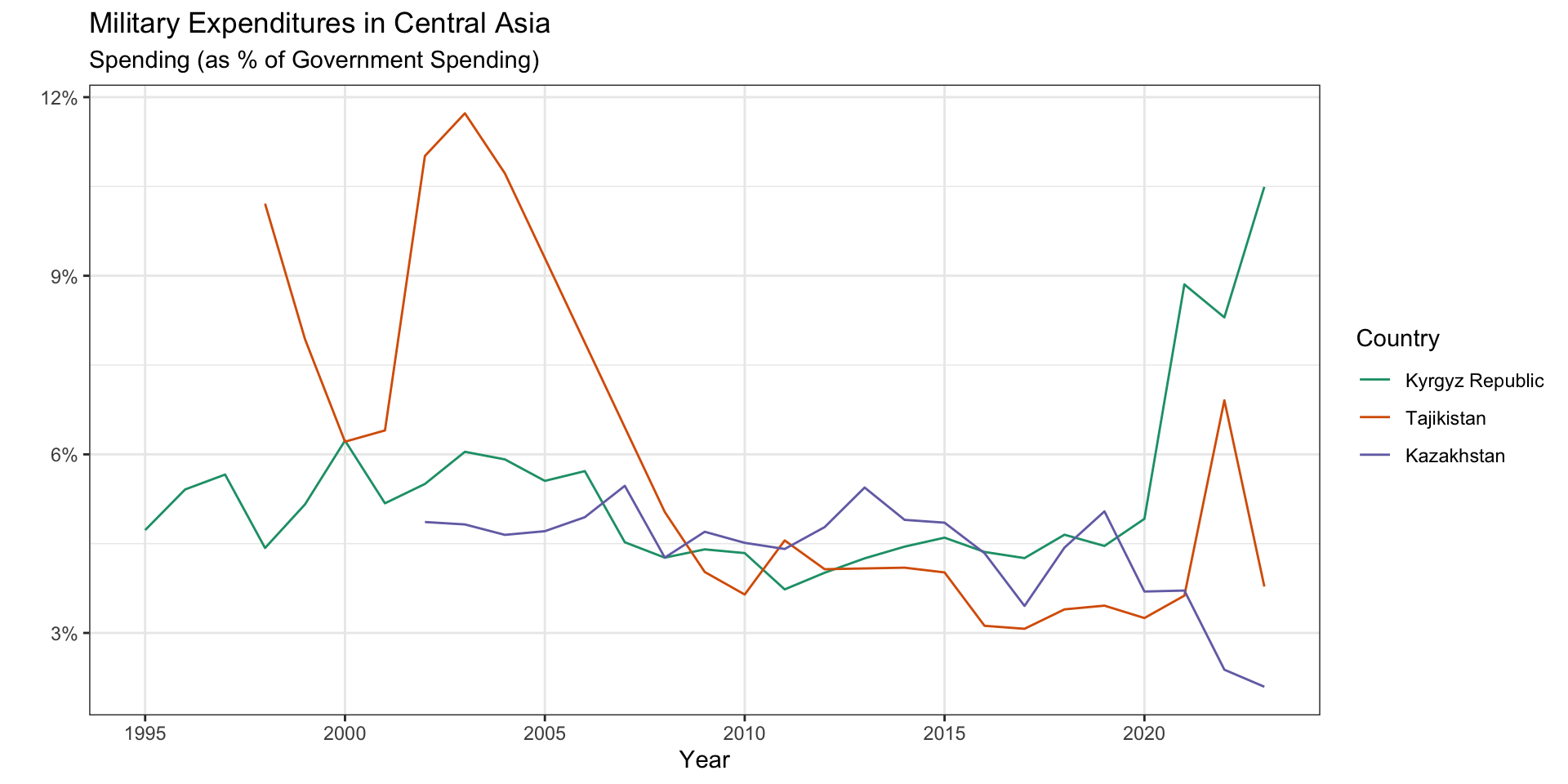
Why Mess with Factors?
Discussion
What are some of the benefits to re-ordering or re-leveling factors variables?
janitor Package
Variable Names in R
Data from external sources likely has variable names not ideally formatted for R.
Names may…
- contain spaces.
- start with numbers.
- start with a mix of capital and lower case letters.
Messy Variable Names are a Pain
You should have noticed this in Practice Activity 4 working with the SIPRI data
You have to use back tick marks around variables that start with numbers or have spaces:
- I personally find capitilization in variable names is very annoying and slows me down
janitor to the rescue!

Mr. Johnson from Abbot Elementary (https://giphy.com/abcnetwork)
Clean Variable Names with janitor
The janitor package converts all variable names in a dataset to snake_case.
Names will…
- start with a lower case letter.
- have spaces and special characters filled in with
_.
Lifecycle Stages
R Is Always Evolving
- Remember:
Ris open source so folks are always adding and updating packages and functions
Discussion
What benefits and drawbacks of R’s ever-evolving nature have you noticed?
- always (aiming) to get better!
- responsive to user input
- new functionality for new statistical methods
- code may no longer run after an update 😭
- need to learn new syntax
- have to keep track of it all 🫠
Lifceycle Stages
As packages get updated, the functions and function arguments included in those packages will change.
- The accepted syntax for a function may change.
- A function/functionality may disappear.

Learn more about lifecycle stages of packages, functions, function arguments in R.
Lifceycle Stages


Deprecated Functions
A deprecated functionality has a better alternative available and is scheduled for removal.
- You get a warning telling you what to use instead.
Warning: Using `across()` in `filter()` was deprecated in dplyr 1.0.8.
ℹ Please use `if_any()` or `if_all()` instead.# A tibble: 3 × 8
Country `1988.0` `1989.0` `1990.0` `1991.0` `1992.0` `1993.0` `1994.0`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 North Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3 sub-Saharan Af… <NA> <NA> <NA> <NA> <NA> <NA> <NA> Deprecated Functions
You should not use deprecated functions!
Instead, we use…
# A tibble: 3 × 8
Country `1988.0` `1989.0` `1990.0` `1991.0` `1992.0` `1993.0` `1994.0`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 North Africa <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3 sub-Saharan Af… <NA> <NA> <NA> <NA> <NA> <NA> <NA> Superceded Functions
A superseded functionality has a better alternative, but is not going away.
- This is a softer alternative to deprecation.
- A superseded function will not give a warning (since there’s no risk if you keep using it), but the documentation will give you a recommendation.
Lab 4: Childcare Costs in California
To do…
- Lab 4: Childcare Costs in California
- Due Monday 4/28 at 11:59pm
- Read Chapter 5: Strings + Dates
- Check-in 5.1 - 5.2 due Tuesday (4/29) before class